Category: technology
Migrated
Goodbye Steve Jobs
My general purpose bioinformatics toolbox
I spend most of my time writing code and using an range of bioinformatics analysis packages. Unlike in many other professions, sometimes there are no best tools for accomplishing a bioinformatics task. The tools are continuously improved and the choice of tools is dependent on the research question and the biology of whatever you are investigating. However I have come to rely on some general purpose resources that make me more productive. Let me introduce you to my general purpose spanner box.
Code Editing
I have finally found nirvana in MacVim, which is the preferred version of Vim for Mac OS. It allows screen splitting, window resizing and integrates with the console, such that you can run system commands right in the editor. You have to install the necessary scripts or plugins to support what you want to do.
It has increased my productivity, although it has a slightly steep learning curve. This is a tool I recommend to any bioinformatician, if you are not already using it!
Cost: Free
Source Control
I use git for source control. It is awesome and fits very well with my workflow. Git has powerful features and easy to use and work with. I like the idea of distributed source control and it makes it easier to work on different versions of the same project!
Cost: Free
Bibliography manager
I use Papers, which is a commercial tool but I would recommend it to anyone. It helps me sort,annotate and read research articles. I once used Mendeley, which is an awesome tool as well.
Cost: $42 (has academic discounts)
Terminal
One of the best tools which we may forget is a tool is the terminal! Since I use Mac OS, I enjoy the best of both worlds, a powerful Unix command line support, excellent graphics and support for proprietary software if need arises.
Pen
I don’t keep an electronic notebook since I prefer jotting down my notes and having a Notebook. I use a liquid ink Pilot V ball RT pen. The pen has a retractable cone-tip liquid ink rollerball, rubber grip and metal pocket clip. It is airplane-safe and writes a 0.4mm line.
Cost: $5
NoteBook.
My preference for a notebook is an A4 D66174 Notebook. Each book has about 180 pages. It comes with a protective handcover. This is an archive for my written thoughts, discussions and workflows.
Cost $90 for a pack of five books.
What is your general purpose bioinformatics toolbox?
My first Bioruby plugin calculates the isoelectric point of a protein
Late last year, there was a lot of talk about creating a plugin system for Bioruby. The idea is that more people can start to develop bioinformatics libraries using the Ruby language and the libraries can leverage on the bioruby framework. Bioruby maintainers can then concentrate on yet to be defined “core” parts of the library to ensure compatibility and support for the plugins.Together with Pascal Bentz we have created a library to calculate the Isoelectric point of a protein given a Pka set and an amino acid sequence of a peptide/protein. The project lay domant for a while at github until now! I am happy to release my first bioruby plugin, bio-isoelectric point! Download it at rubygems.org Fork it and check the usage at github
Examples
require 'bio'
require 'bio-isoelectric_point'
protein_seq = Bio::Sequence::AA.new("KKGFTCGELA")
#what is the protein charge at ph 14?
charge = protein_seq.charge_at(14) #=>-2.999795857467562
#calculate the ph using dtaselect pka set and round off to 3 decimal places
isoelectric_point = protein_seq.isoelectric_point(‘dtaselect’, 3) #=>8.219
# calculate the isoelectric point pH with a custom set
custom_pka_set = { “N_TERMINUS” => 8.1,
“K” => 10.1,
“R” => 12.1,
“H” => 6.4,
“C_TERMINUS” => 3.15,
“D” => 4.34,
“E” => 4.33,
“C” => 8.33,
“Y” => 9.5
}
iep_ph = protein_seq.isoelectric_point(custom_pka_set, 3) #=> 8.193
This gem supports the following Pka sets, as well as allowing a user to provide a custom Pka set.
* dta_select
* emboss
* rodwell
* wikipedia
* sillero
Happy biology!
PC vs Apple Mac (Not the war!)
My good old PC running Linux OS is coming of age and recently started failing. The Optical drive is not functional and occasionally it will freeze. The Top cover does not hold anymore and the graphical TFT screen needs to be supported carefully.
While this particular computer has served me well, I am at that point where i need a new machine but am torn between an Apple Mac and a PC running Linux. First my work involves the following aspects;
- Compiling and running bioinformatics software developed using open source standards and technologies
- Programming
- Word processing and document editing
- Occasional mathematical modeling
- Administering Unix based servers
I have tried to come up with a computer-model agnostic specifications for my needs.
Hardware
* High Processor speed (2.60GHZ or above)
* High Memory (4GB or above)
* Medium Hard-disk space (160GB and above)
* Long Battery life ( 5hours and above)
* Durable external cover
* Ergonomic keys and mouse
* Support for multiple external devices(printers,Cameras,Microphones,Storage devices,monitors)
* Excellent support for wireless technologies
* Support for running multiple operating systems on the same machine
Size and weight
* Lightweight
* convenience while travelling while traveling
Operating System
* A Unix or Linux derived operating system
* Easy to upgrade at zero or minimal cost
* Free patches against known security holes and problems.
Software
* Support for Open software standards
* Support for Microsoft, Adobe and other proprietary software vendor’s products
Security
* Excellent inbuilt support against Malware, Trojans and viruses at minimal or no cost
* Support for locking the machine while away or against unauthorized login
* Ability to easily ‘tag’ the machine in case of theft
Price : Affordable and reasonable
Based on the above specifications I have evaluated two computers models that can satisfy the above needs.
1. A PC laptop computer running a Linux based operating system
2. An Apple Macintosh laptop computer
I have ruled out a Windows/DOS based Operating software because Microsoft Windows based operating system cannot offer support for open source standards and technologies. OS upgrade for windows is very expensive and the OS is highly prone to malware, Trojans and viruses. Most bioinformatics software and tools are developed on Unix or Linux environment.
PC can support Linux installations even though one looses on hardware optimization. Linux has a relatively poor graphical user interface and functionality when compared to Mac OS or Windows. There is limited support for document processing, graphics and rich multimedia applications support. Linux does not support any of the Microsoft software applications natively. There are open source equivalents but most lack good support.
Apple Macintosh computers are based on Unix and open source technologies, they support both closed source and open source standards. The hardware is optimized and accelerated for the Apple mac OS. They offer excellent graphical user interface system, a powerful terminal for interaction with the OS, they are not prone to virus attacks, and they support long battery life as well as portability, ergonomics and a relatively within a price range equivalent to a PC of the same specifications.
Given my budget constrains, I am thinking that a 2.53GHz Apple Macintosh 13 inch model with 4GB of memory is best for my needs. There is little price differences between the PC and Macintosh models based on my specifications. PC models do not favor Linux installations and Linux hardware support is not guaranteed. They however seem to have a more flexible price ranges depending on the manufacturers, vendors, quality and specifications.
I will keep Linux to run my server applications.
Bio-graphics, BioSQL and Rails part 2
In part 1 of this series we created a rails application and connected it to a BioSQL database. We also overwrote the rails convections to accommodate our legacy schema.
To understand the BioSQL schema, please review the documentation here. A brief overview of is as follows. Every record we enter into our database is a ‘bioentry’ and goes to the bioenty table. A bioentry can be composed of the following entities: the record’s public name, public accession and version, its description and an identifier field.
The actual sequence data is stored in the biosequence table which contains raw sequence information associated with a bioentry, and alphabet information (‘protein’, ‘dna’, ‘rna’). This is because not all records in our database need to be associated with a raw sequence. Additional sequence information is stored in the seqfeature table together with other qualifiers.
The location of each seqfeature (or sub-seqfeature) is defined by a location entity, describing the stop and start coordinates and strand. This information is stored in the location table.
In our rails application we are going to create some models and a few controllers. In RESTful language, we are actually creating resources. In this example we will be very simplistic and just create a biodatabase, taxon, bioentry, biosequence, seqfeature, location resources. We will also create associations between them in their model classes. But before that delete the index.html file from your rails application public folder and add the following line to your configurations/routes.rb file
map.root :controller => "biosequences"
To quickly create the models, controllers, associated views and a test suite for each of our resources, just run the rails generate scaffold command, passing the name of the model as an argument. For example,
generate scaffold Bioentry
will create a bioentry model, a bioentries_controller, associated views (index,show,edit and new), a migration file, though in our case we do not need it. When you finish scaffolding, the routes.rb file should have the following resources declared.
map.resources :seqfeatures
map.resources :locations
map.resources :bioentries
map.resources :biosequences
map.resources :taxon
map.resources :biodatabases
Let us create some mandatory associations for the models.
Edit the /models/biodatabase.rb file by adding the following
has_many :bioentries #a biodatabase is associated with many bioentries
validates_uniqueness_of :name #The name foe each biodatabase is unique!
Edit the /models/bioentry.rb file by adding the following
belongs_to :biodatabase
belongs_to :taxon
has_one :biosequence
Edit the /models/taxon.rb and add
has_one :bioentry
Edit the /models/biosequence.rb file by adding:
set_primary_key :bioentry_id #biosequence uses bioentry_id as a primary key! belongs_to :bioentry
edit the /models/location.rb file by adding:
belongs_to :seqfeature
Edit the /models/seqfeature.rb file by adding:
belongs_to :bioentry
has_many :locations
Note that most likely you will be adding huge files to the database. BioSQL comes with a set of perl scripts to enable you do that. Until bioruby 1.3 is released you will have to use the perl scripts to add huge datasets. All the documentation to do that is available from the BioSQL website. I used a perl script load_ncbi_taxonomy.pl to load taxon data to my database. This script comes with the BioSQL. (It did not seem to work on my system, I will sort that later)
To make this post shorter and get to the meat of it, i will assume that you have some existing data in your biosql database. If not, create some dummy data to populate, the biodatabase, bioentry,biosequence, seqfeature and location tables. In Part 3, I will show you how to create the necessary views to populate the database. After all biologists don’t want to interact with raw SQL queries and sometimes have no idea of running scripts, however they are very web savy!
Edit the /biosequences/show.html.erb to look as follows:
<h2><%= @biosequence.bioentry.name%>(<%= @biosequence.alphabet %>)</h2> <p>Sequence</p> <%= @biosequence.seq %><br/> <%= link_to 'Edit', edit_biosequence_path(@biosequence) %>
Now navigate to http://localhost:3000/biosequences/1
and then navigate to http://locahost:3000/biosequences/1.xml The XML version of your sequence is also available!
Lets add some ability to render graphics for the sequences.
Add the following lines at the top of the biosequence.rb model file
require 'stringio'
require 'base64'
In the biosequence.rb model class, create a new method called draw_graphic.
def self.draw_graphic(value) #get the name and length of the main feature to be drawn main_feature = Bioentry.find(value) len = main_feature.biosequence.length.to_i name = main_feature.name #create a Biographics panel and add a track @my_panel = Bio::Graphics::Panel.new(len,:width=> 900) @track = @my_panel.add_track("#{name}",:glyph=>'directed_generic') #specify the range for the main feature main_feature_range = "1..#{len}" @track.add_feature(Bio::Feature.new("#{name}",main_feature_range), :label=>" ") #write the output to memory output = StringIO.new @my_panel.draw(output) return output.string end
This method will be called by an action method in biosequence_controller.rb file.
def to_image
begin
image = Biosequence.draw_graphic(Biosequence.find(params[:id]))
send_data(image, :filename => "graphic.svg", :disposition => "inline")
rescue ActiveRecord::RecordNotFound
add_error("Error:Attempt to call image without specifying a biosequence ID")
redirect_to :action=>'index'
end
end
We add a rescue block to capture record not found errors. In RESTful applications a controller is limited to seven actions. So we need to add a collection to our biosequence resource in routes.rb. This is how we do it.
map.resources :biosequences,:collection=>{:to_image=>:get}
Now we need to modify our /biosequences/show.html.erb file, to enable rendering of the graphic. For that we will create a helper method so that our show.html.erb view is ‘clean’. In helpers/biosequences_helper.rb file, add the following code
def render_image(feature_obj)
image_tag(url_for({:action=>'to_image',:id=>feature_obj}))
end
And in the /views/biosequences/show.html.erb file add the following line of code
<%= render_image(@biosequence) %><br/>
Now assuming that you have a biosql database with valid data, navigate to
http://localhost:3000/biosequences/show/1
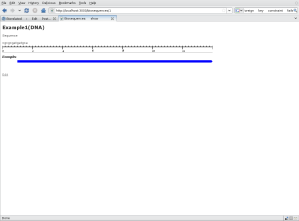
screenshort
The above is a screen shot from my example application while I was writing this tutorial.
The source code for this example application is available from github
For a full review of the methods available for biographics please check the project’s git repository and the rdoc.
At the Bench Series
Am cross cutting between the bench and the computer. My mentor and supervisor has given me an excellent book called At the Bench: A laboratory Navigator by Kathy Barker. It is an exciting read for anyone who plans to use a biomedical research laboratory and I thought, Why not share some of the gems as I read along:
Basic Survival
Simple lab courtesy is a nice way of maintaining healthy working relationship with your fellow lab rats, in this rat race! What Kathy says sounds real trouble but i think and you will probably agree, its basic common sense.
Attitude
- Ask, do not command.
- Assume nothing
- Write down everything when given instructions
- Make appointments or request time with people
- Do not remove journals from the Departmental library
- Do not discuss a fellow lab rat’s results with people not in the lab
Courtesy at the Bench
- Never use reagents or buffers without permission
- Do not ignore broken equipments
- Order common reagents if they are running low
- Do not move things around or change locations of any tubes reagents or equipment.(This is not ya damn bedroom!)
- Do not leave anything anywhere
- If you do something wrong, confess (I wonder whether there is purgatory :))
- Clean up immediately after an experiment
- Request the minimum of favors( fellow lab rats are not working for you)
Like all complex organizations, each research lab has its own culture and rules. the rules are largely unspoken and may not be written down. You are expected to know how to work with the equipment or referred to the manual. You are required to decipher the vague signs and complex language to understand the rhythm and beat of the lab. Kathy does a great job in introducing the naive to the workbench in this book.
All the best dear friend.
A ruby micro review
 Ruby is a reflective, dynamic, object-oriented programming language, created by Yukihiro Matsumoto and released to the public in 1995. It is an extremely pragmatic language, less concerned with formalities and more concerned with ease of development and valid results. You will see Agile principles running through ruby and particularly with rails. Most of all TDD and BDD concepts / philosophies have been implemented for ruby developers. Ruby differs from most programming languages by syntax, culture, grammar and customs. It has more in common with LISP and Smalltalk than with most languages such as C++ and PHP.
Ruby is a reflective, dynamic, object-oriented programming language, created by Yukihiro Matsumoto and released to the public in 1995. It is an extremely pragmatic language, less concerned with formalities and more concerned with ease of development and valid results. You will see Agile principles running through ruby and particularly with rails. Most of all TDD and BDD concepts / philosophies have been implemented for ruby developers. Ruby differs from most programming languages by syntax, culture, grammar and customs. It has more in common with LISP and Smalltalk than with most languages such as C++ and PHP.
If you can program in languages such as Perl, PHP, C or Pascal, using and learning ruby is quite easy, but the problem solving pespectives that ruby uses may throw you out at first.
The so popular and hyped ruby on rails DSL (domain specific language) is a framework for developing web applications and currently powers hundreds of large websites around the world.
Bioruby is an excellent bioinformatics library for ruby. Though not highly documented like its sister, bioperl, efforts are been made to improve its level of documentation. The bioruby community is also really nice and friendly. Not a single question that i have posted on the mailing list goes unanswered.
Hundreds of libraries for performing different tasks have been written for ruby , packaged as gems and hosted at rubyforge
So far my favorite ruby editor is the netbeans IDE, whose currently release is in beta 2. The final release is slated for 3rd of Dec 2007. (Am waiting!). It features auto completion, syntax highlighting among other cool things that makes programming a joy. It also comes bundled with the jruby release, a java implementation of ruby that is starting to rock the world, so you can choose to use either native ruby or jruby, the choice is all yours!
Ruby can be downloaded here
Standalone BLAST with Ruby (windows)
#create a query sequence
myseq="pcaatcacatyyawwqqffgghhhkllkl"
#create a temporary file
require 'tempfile'
temp=Tempfile.new("seqfile")
#get the name of the temporary file
name=temp.path
#append the contents your sequence to this temporary file
temp.puts "#{myseq}"
temp.close
#since we have a protein query sequence, we will run a blastp. Please note that you will need to have a valid #database to query against. use the formatdb command to create your database before executing the lines #below.
@program = 'blastp'
#path to blast
@database = 'c:/path_to_databasefile'
#name of your query file
@input= name
#your blast output file
@output='c:/path_output_file'
#assume your blast is in a folder called NCBI_Blast, execute
system( "c:/NCBI_Blast/bin/blastall.exe -p #{@program} -d #{@database} -i #{@input} -o #{@output}")
#To capture the output in a variable execute this command instead.
#note that we have omitted the blast -o parameter
result=%x(c:/NCBI_Blast/bin/blastall.exe -p #{@program} -d #{@database} -i #{@input} )
#remember to delete the temporary file!
temp.close(true)