Bio-graphics, BioSQL and Rails part 2
In part 1 of this series we created a rails application and connected it to a BioSQL database. We also overwrote the rails convections to accommodate our legacy schema.
To understand the BioSQL schema, please review the documentation here. A brief overview of is as follows. Every record we enter into our database is a ‘bioentry’ and goes to the bioenty table. A bioentry can be composed of the following entities: the record’s public name, public accession and version, its description and an identifier field.
The actual sequence data is stored in the biosequence table which contains raw sequence information associated with a bioentry, and alphabet information (‘protein’, ‘dna’, ‘rna’). This is because not all records in our database need to be associated with a raw sequence. Additional sequence information is stored in the seqfeature table together with other qualifiers.
The location of each seqfeature (or sub-seqfeature) is defined by a location entity, describing the stop and start coordinates and strand. This information is stored in the location table.
In our rails application we are going to create some models and a few controllers. In RESTful language, we are actually creating resources. In this example we will be very simplistic and just create a biodatabase, taxon, bioentry, biosequence, seqfeature, location resources. We will also create associations between them in their model classes. But before that delete the index.html file from your rails application public folder and add the following line to your configurations/routes.rb file
map.root :controller => "biosequences"
To quickly create the models, controllers, associated views and a test suite for each of our resources, just run the rails generate scaffold command, passing the name of the model as an argument. For example,
generate scaffold Bioentry
will create a bioentry model, a bioentries_controller, associated views (index,show,edit and new), a migration file, though in our case we do not need it. When you finish scaffolding, the routes.rb file should have the following resources declared.
map.resources :seqfeatures
map.resources :locations
map.resources :bioentries
map.resources :biosequences
map.resources :taxon
map.resources :biodatabases
Let us create some mandatory associations for the models.
Edit the /models/biodatabase.rb file by adding the following
has_many :bioentries #a biodatabase is associated with many bioentries
validates_uniqueness_of :name #The name foe each biodatabase is unique!
Edit the /models/bioentry.rb file by adding the following
belongs_to :biodatabase
belongs_to :taxon
has_one :biosequence
Edit the /models/taxon.rb and add
has_one :bioentry
Edit the /models/biosequence.rb file by adding:
set_primary_key :bioentry_id #biosequence uses bioentry_id as a primary key! belongs_to :bioentry
edit the /models/location.rb file by adding:
belongs_to :seqfeature
Edit the /models/seqfeature.rb file by adding:
belongs_to :bioentry
has_many :locations
Note that most likely you will be adding huge files to the database. BioSQL comes with a set of perl scripts to enable you do that. Until bioruby 1.3 is released you will have to use the perl scripts to add huge datasets. All the documentation to do that is available from the BioSQL website. I used a perl script load_ncbi_taxonomy.pl to load taxon data to my database. This script comes with the BioSQL. (It did not seem to work on my system, I will sort that later)
To make this post shorter and get to the meat of it, i will assume that you have some existing data in your biosql database. If not, create some dummy data to populate, the biodatabase, bioentry,biosequence, seqfeature and location tables. In Part 3, I will show you how to create the necessary views to populate the database. After all biologists don’t want to interact with raw SQL queries and sometimes have no idea of running scripts, however they are very web savy!
Edit the /biosequences/show.html.erb to look as follows:
<h2><%= @biosequence.bioentry.name%>(<%= @biosequence.alphabet %>)</h2> <p>Sequence</p> <%= @biosequence.seq %><br/> <%= link_to 'Edit', edit_biosequence_path(@biosequence) %>
Now navigate to http://localhost:3000/biosequences/1
and then navigate to http://locahost:3000/biosequences/1.xml The XML version of your sequence is also available!
Lets add some ability to render graphics for the sequences.
Add the following lines at the top of the biosequence.rb model file
require 'stringio'
require 'base64'
In the biosequence.rb model class, create a new method called draw_graphic.
def self.draw_graphic(value) #get the name and length of the main feature to be drawn main_feature = Bioentry.find(value) len = main_feature.biosequence.length.to_i name = main_feature.name #create a Biographics panel and add a track @my_panel = Bio::Graphics::Panel.new(len,:width=> 900) @track = @my_panel.add_track("#{name}",:glyph=>'directed_generic') #specify the range for the main feature main_feature_range = "1..#{len}" @track.add_feature(Bio::Feature.new("#{name}",main_feature_range), :label=>" ") #write the output to memory output = StringIO.new @my_panel.draw(output) return output.string end
This method will be called by an action method in biosequence_controller.rb file.
def to_image
begin
image = Biosequence.draw_graphic(Biosequence.find(params[:id]))
send_data(image, :filename => "graphic.svg", :disposition => "inline")
rescue ActiveRecord::RecordNotFound
add_error("Error:Attempt to call image without specifying a biosequence ID")
redirect_to :action=>'index'
end
end
We add a rescue block to capture record not found errors. In RESTful applications a controller is limited to seven actions. So we need to add a collection to our biosequence resource in routes.rb. This is how we do it.
map.resources :biosequences,:collection=>{:to_image=>:get}
Now we need to modify our /biosequences/show.html.erb file, to enable rendering of the graphic. For that we will create a helper method so that our show.html.erb view is ‘clean’. In helpers/biosequences_helper.rb file, add the following code
def render_image(feature_obj)
image_tag(url_for({:action=>'to_image',:id=>feature_obj}))
end
And in the /views/biosequences/show.html.erb file add the following line of code
<%= render_image(@biosequence) %><br/>
Now assuming that you have a biosql database with valid data, navigate to
http://localhost:3000/biosequences/show/1
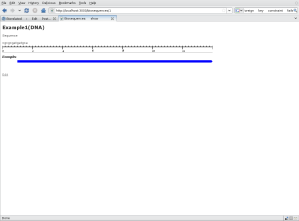
screenshort
The above is a screen shot from my example application while I was writing this tutorial.
The source code for this example application is available from github
For a full review of the methods available for biographics please check the project’s git repository and the rdoc.
This is a very useful, helpful series – thanks!
Noticed that part of your code (biosequence.rb, above) has converted to an emoticon. I guess it is:
Bio::Graphics::Panel.new
There’s a WordPress config option to stop this – don’t recall where just now.
Oh look, my comment has the same problem.
Thank you Saunders!
I think it has to do with my wordpress blog settings. I will disable the emotions if the settings are available. That line should read Bio::Graphics(double colon)Panel.new
Just dropping by.Btw, you website have great content!
______________________________
Instantly Search Millions of Public Records & Resources
good job on the part 1 and part 2…. it helps a lot for my understanding on biographic, bioSQL…
Superb website…
Any new update on Part 3…? it has been 5 months since part 2….Looking forward for ur update…
Thanks.
Excellent blog! Very interesting themes. I will regularly read it.
Hi George,
My name is Darren. I’m the ex-technology editor at BBC News and I’m now working with Mendeley.
I saw your Tweet about Mendeley and I wondered if I could speak with you about possibly doing an interview with the BBC World Service about open science and how tools like Mendeley are driving collaboration.
Do you have an email address or telephone number/Skype that we could chat on?
thanks.
Darren
Hey there, I’ve been reading your weblog for about a month now. So I just decided to stop lurking and say hi :)
Great, thanks. One of the better things I’ve read today! Was actually looking for it for the last few hours.